Dartmon's BRAF antibody firmly underlies melanoma diagnosis and treatment
Issuing time 2025-04-14 15:43:07
BRAF is a serine/threonine protein kinase, encoded on chromosome 7q34, that activates the MAP kinase/ERK-signaling pathway. BRAF is the family member most easily activated by Ras.
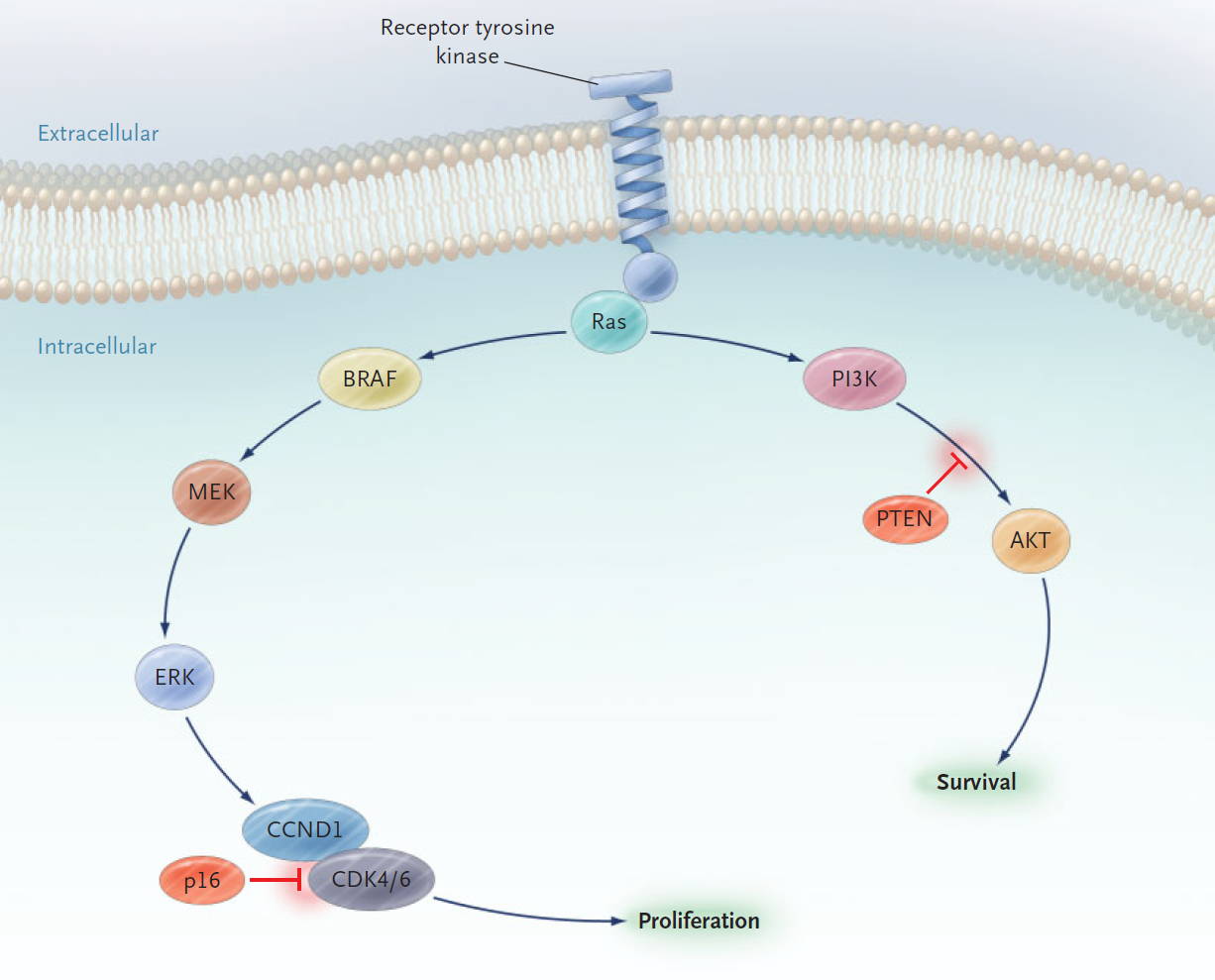
Figure 1. The Mitogen-Activated Protein (MAP) Kinase and Phosphatidylinositol 3' Kinase (PI3K) Pathways. Signals from receptor tyrosine kinases can promote proliferation through the MAP kinase pathway (left branch) and survival through the PI3 kinase pathway (right branch).
In 2002, Davies H, et al. first reported that BRAF mutations occur at a high frequency in various cancers (melanoma, thyroid cancer, lung adenocarcinoma, colorectal cancer) and demonstrated its oncogenicity. [1]
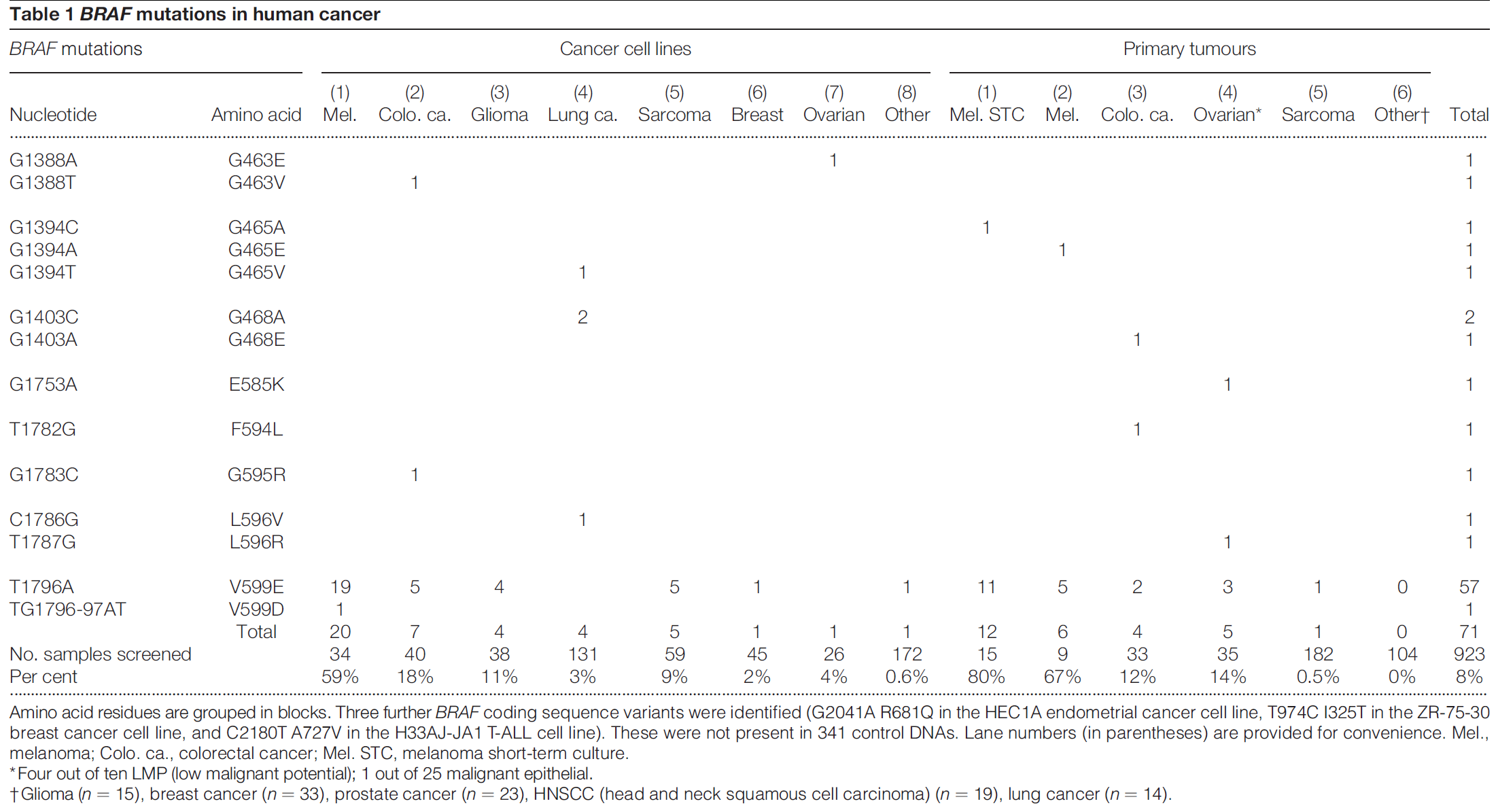
In 2005, Curtin JA, et al. revealed the distribution of BRAF mutations in melanoma and the relationship between these mutations and ultraviolet (UV) exposure. [2]
Oncogenic activation of the MAPK pathway can occur via multiple mechanisms, the most common of which in melanoma is constitutive activation of the BRAF kinase via mutation, which occurs in ~ 40–60% of cases. The second most common MAPK pathway aberration in melanoma is mutated NRAS, occurring in~ 15–30% of cases. [3-7]
BRAF encodes a cytoplasmic serine–threonine kinase. More than 97% of BRAF mutations are located in codon 600 of the BRAF gene. [8] Types and frequencies of BRAF mutations are summarized in following table and picture. [9,10]
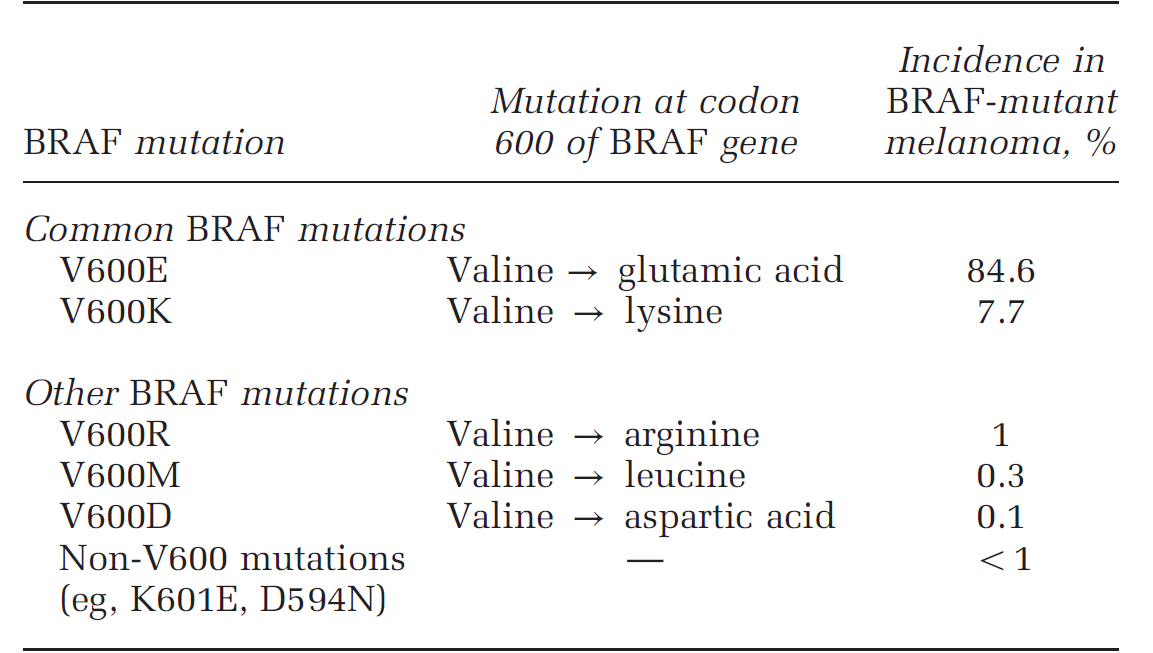
Table 2. Incidence of common BRAF mutations based on the COSMIC database
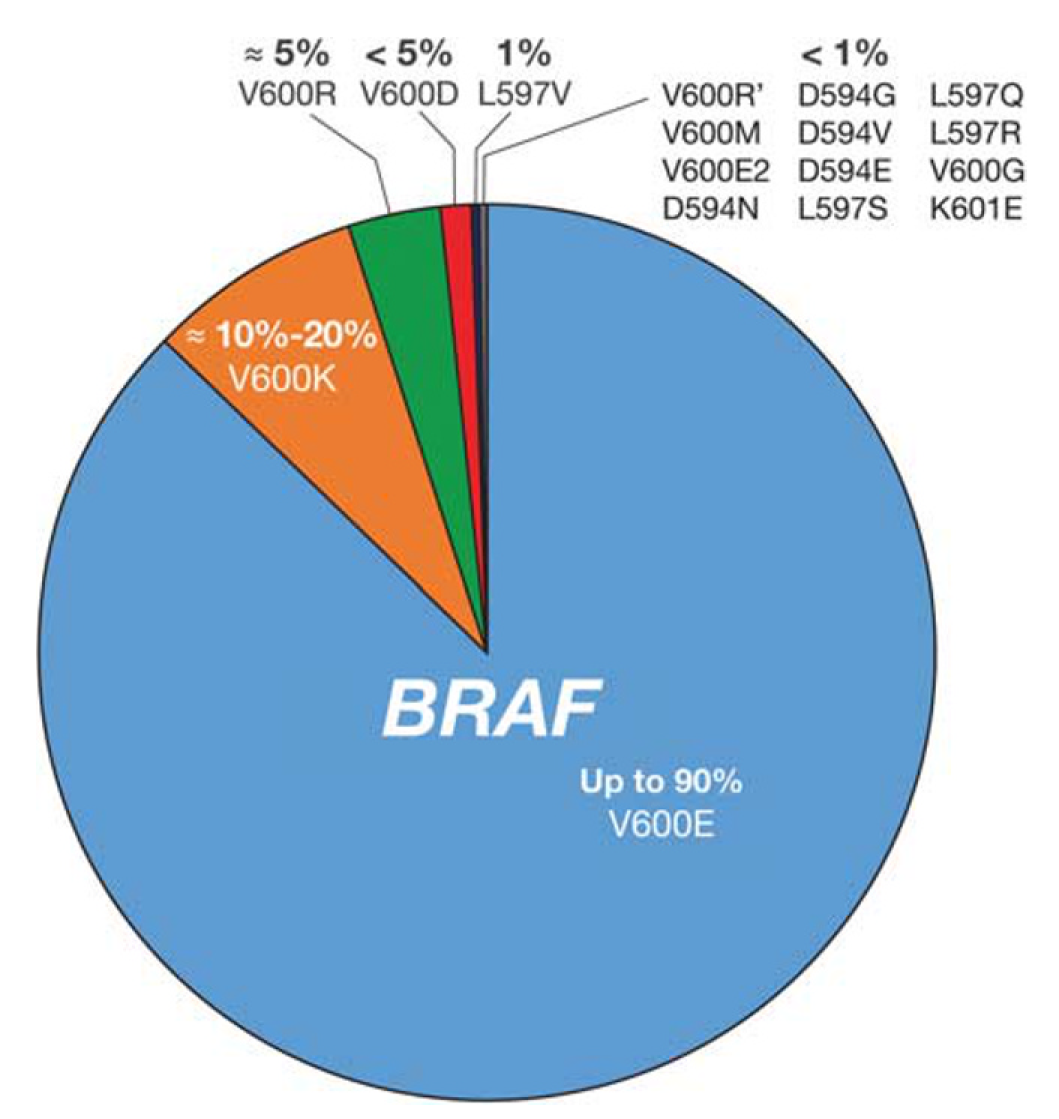
Figure 2. BRAF mutation types in melanoma. Estimated incidence of BRAF mutation frequencies in patients with melanoma is shown.
It is of paramount importance that a patient’s BRAF mutational status is promptly and accurately determined at the time of initial diagnosis, because it is currently the only reliable predictive biomarker that can influence the treatment of advanced melanoma. Immunostaining for the detection of BRAF-mutated protein is a quick and inexpensive test that can be combined with other melanoma markers.
For melanoma tissues, the immunohistochemistry staining should be carefully evaluated, as melanin has a color similar to that of DAB, making it difficult to distinguish between them, which may lead to misjudgment. We recommend setting up blank controls or using AP chromogenic agents for color development.
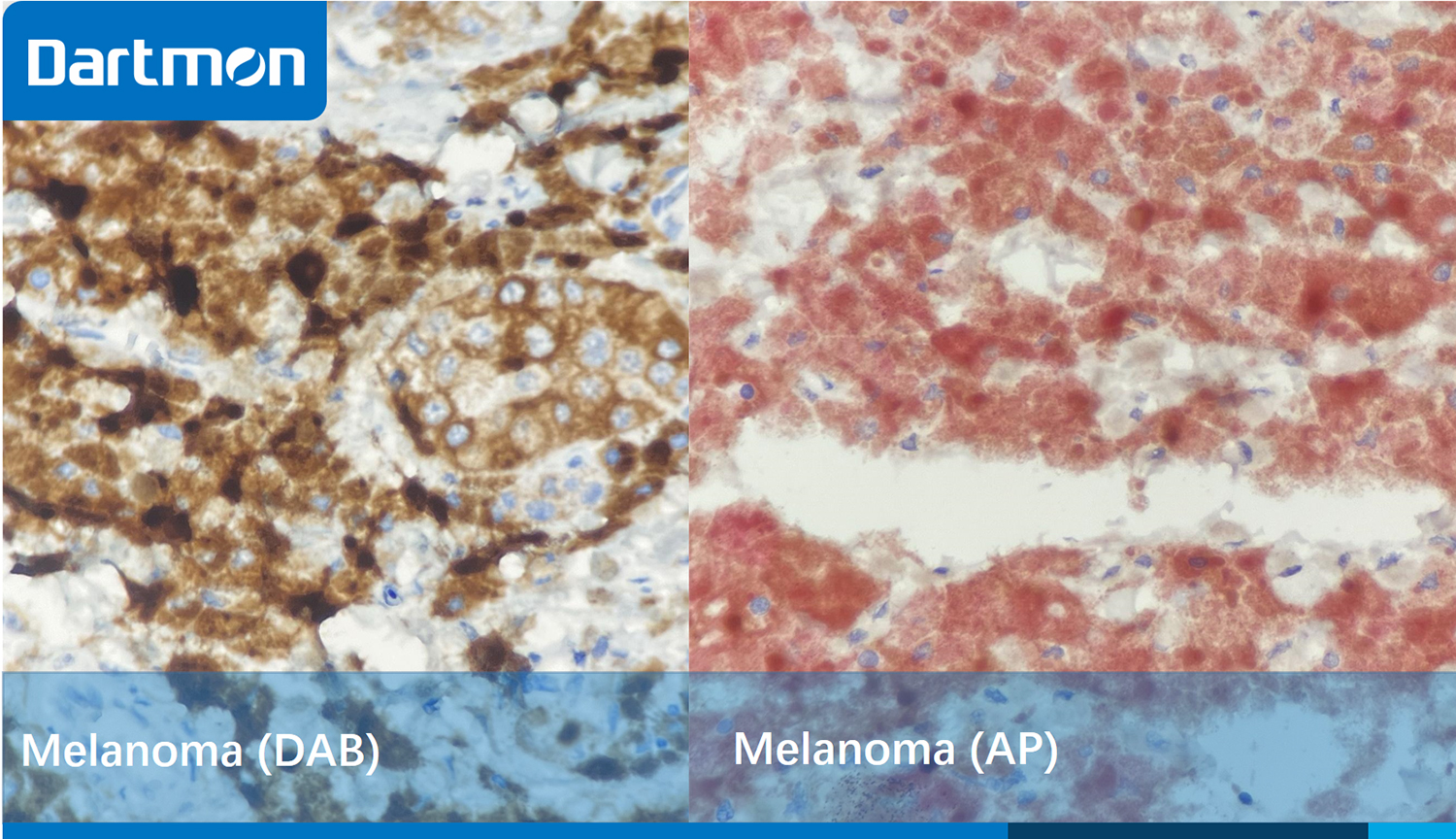
Figure 3. The picture on the left depicts a malignant melanoma of the intestine, which was stained with Antibody BRAF and DAB. Due to the similarity between the color of melanin and the brownish-yellow color of DAB, it is difficult to distinguish between melanin and the positive localization. The picture on the right shows the same tissue, which was stained with Antibody BRAF and AP. It is clearly visible that there is a strongly positive expression, with the cytoplasm of the tumor cells showing a red color.

Figure 4. Serrated adenoma of the intestine. BRAF protein is located in the cytoplasm of tumor cells, showing fine granular cytoplasmic staining.

Figure 5. Thyroid cancer. The tumor cells show moderate to strong positive staining. Due to the tumor heterogeneity, the content of BRAF protein expressed by different tumor cells is not the same.

Figure 6. Papillary thyroid carcinoma. Due to tumor heterogeneity, the content of BRAF protein expressed by different tumor cells varies.

Product name: Anti- BRAF (DA007)
Cat. No. : RMB1A106
Usage pattern: Manual or device utilization
Ready-to-use: 3ml, 6ml, 10ml
Concentrated: 0.1ml, 0.5 ml, 1ml


References
[1] Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N. Mutations of the BRAF gene in human cancer. Nature. 2002 Jun 27;417(6892):949-54.
[2] Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Bröcker EB, LeBoit PE, Pinkel D. Distinct sets of genetic alterations in melanoma. New England Journal of Medicine. 2005 Nov 17;353(20):2135-47.
[3] Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, Massi D, Fonsatti E, Staibano S, Nappi O, Pagani E. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. Journal of Clinical Oncology. 2012 Jul 10;30(20):2522-9.
[4] Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H, Jakrot V. Whole-genome landscapes of major melanoma subtypes. Nature. 2017 May 11;545(7653):175-80.
[5] Fedorenko IV, Gibney GT, Smalley KS. NRAS mutant melanoma: biological behavior and future strategies for therapeutic management. Oncogene. 2013 Jun;32(25):3009-18.
[6] Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, Hughes TM, Thompson JF, Scolyer RA, Kefford RF. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. Journal of Clinical Oncology. 2011 Apr 1;29(10):1239-46.
[7] Ribas A, Flaherty KT. BRAF targeted therapy changes the treatment paradigm in melanoma. Nature reviews Clinical oncology. 2011 Jul;8(7):426-33.
[8] Ihle MA, Fassunke J, König K, Grünewald I, Schlaak M, Kreuzberg N, Tietze L, Schildhaus HU, Büttner R, Merkelbach-Bruse S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p. V600E and non-p. V600E BRAF mutations. BMC cancer. 2014 Dec;14:1-3.
[9] Bradish JR, Cheng L. Molecular pathology of malignant melanoma: changing the clinical practice paradigm toward a personalized approach. Human Pathology. 2014 Jul 1;45(7):1315-26.
[10] COSMIC. Catalogue of Somatic Mutations in Cancer. Available from http://cancer.sanger.ac.uk/cosmic


